Some years in the past, when working as a marketing consultant, I used to be deriving a comparatively complicated ML algorithm, and was confronted with the problem of constructing the internal workings of that algorithm clear to my stakeholders. That’s after I first got here to make use of parallel coordinates – as a result of visualizing the relationships between two, three, perhaps 4 or 5 variables is simple. However as quickly as you begin working with vectors of upper dimension (say, 13, for instance), the human thoughts oftentimes is unable to know this complexity. Enter parallel coordinates: a software so easy, but so efficient, that I usually marvel why it’s so little in use in on a regular basis EDA (my groups are an exception). Therefore, on this article, I’ll share with you the advantages of parallel coordinates primarily based on the Wine Dataset, highlighting how this system might help uncover correlations, patterns, or clusters within the knowledge with out dropping the semantics of options (e.g., in PCA).
What are Parallel Coordinates
Parallel coordinates are a standard methodology of visualizing high-dimensional datasets. And sure, that’s technically right, though this definition doesn’t totally seize the effectivity and magnificence of the strategy. Not like in a normal plot, the place you have got two orthogonal axes (and therefore two dimensions which you could plot), in parallel coordinates, you have got as many vertical axes as you have got dimensions in your dataset. This implies an commentary may be displayed as a line that crosses all axes at its corresponding worth. Need to study a elaborate phrase to impress on the subsequent hackathon? “Polyline”, that’s the right time period for it. And patterns then seem as bundles of polylines with related behaviour. Or, extra particularly: clusters seem as bundles, whereas correlations seem as trajectories with constant slopes throughout adjoining axes.
Marvel why not simply do PCA (Principal Part Evaluation)? In parallel coordinates, we retain all the unique options, that means we don’t condense the data and undertaking it right into a lower-dimensional house. So this eases interpretation loads, each for you and on your stakeholders! However (sure, over all the thrill, there should nonetheless be a however…) it is best to take excellent care to not fall into the overplotting-trap. In case you don’t put together the info fastidiously, your parallel coordinates simply turn out to be unreadable – I’ll present you within the walkthrough that function choice, scaling, and transparency changes may be of nice assist.
Btw. I ought to point out Prof. Alfred Inselberg right here. I had the honour to dine with him in 2018 in Berlin. He’s the one who bought me hooked on parallel coordinates. And he’s additionally the godfather of parallel coordinates, proving their worth in a mess of use circumstances within the Nineteen Eighties.
Proving my Level with the Wine Dataset
For this demo, I selected the Wine Dataset. Why? First, I like wine. Second, I requested ChatGPT for a public dataset that’s related in construction to considered one of my firm’s datasets I’m at present engaged on (and I didn’t wish to tackle all the effort to publish/anonymize/… firm knowledge). Third, this dataset is well-researched in lots of ML and Analytics purposes. It incorporates knowledge from the evaluation of 178 wines grown by three grape cultivars in the identical area of Italy. Every commentary has 13 steady attributes (suppose alcohol, flavonoid focus, proline content material, color depth,…). And the goal variable is the category of the grape.
So that you can comply with via, let me present you how one can load the dataset in Python.
import pandas as pd
# Load Wine dataset from UCI
uci_url = "https://archive.ics.uci.edu/ml/machine-learning-databases/wine/wine.knowledge"
# Outline column names primarily based on the wine.names file
col_names = [
"Class", "Alcohol", "Malic_Acid", "Ash", "Alcalinity_of_Ash", "Magnesium",
"Total_Phenols", "Flavanoids", "Nonflavanoid_Phenols", "Proanthocyanins",
"Color_Intensity", "Hue", "OD280/OD315", "Proline"
]
# Load the dataset
df = pd.read_csv(uci_url, header=None, names=col_names)
df.head()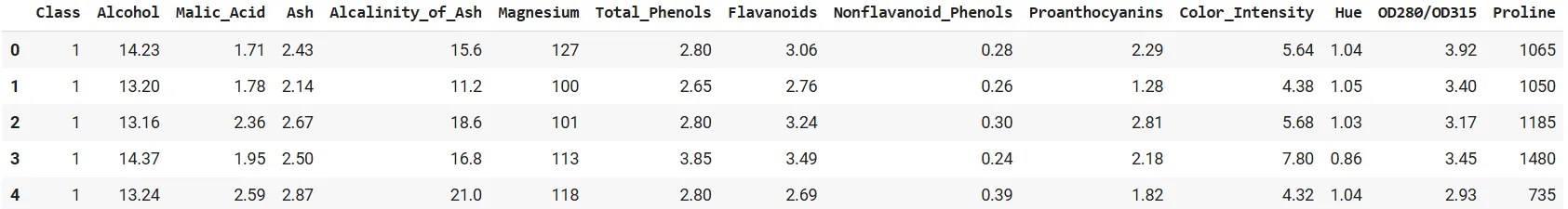
Good. Now, let’s derive a naïve plot as a baseline.
First Step: Constructed-In Pandas
Let’s use the built-in pandas plotting operate:
from pandas.plotting import parallel_coordinates
import matplotlib.pyplot as plt
plt.determine(figsize=(12,6))
parallel_coordinates(df, 'Class', colormap='viridis')
plt.title("Parallel Coordinates Plot of Wine Dataset (Unscaled)")
plt.xticks(rotation=45)
plt.present()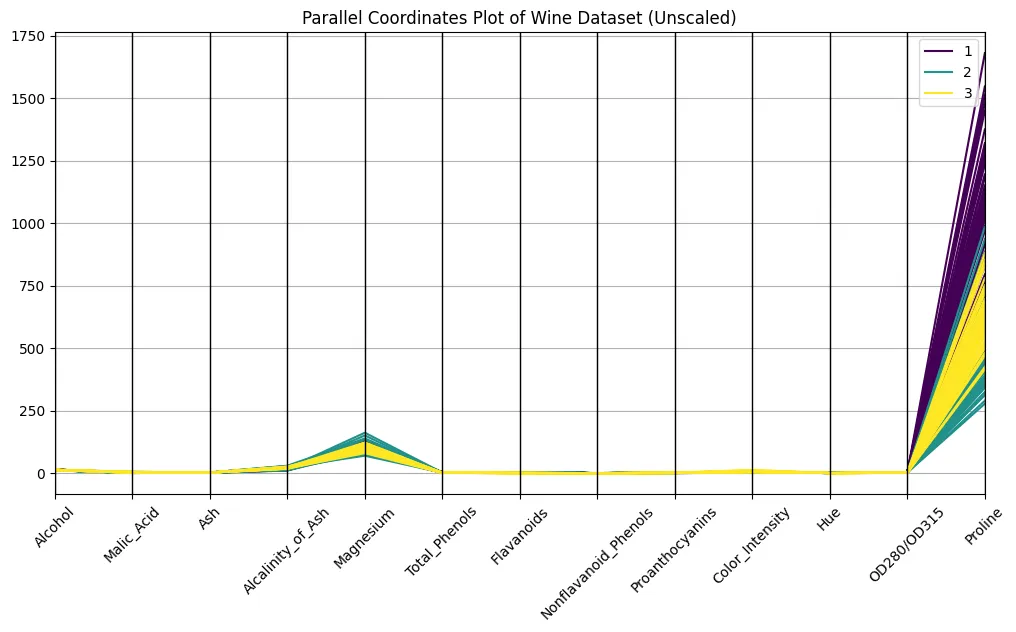
Seems to be good, proper?
No, it doesn’t. You definitely are in a position to discern the lessons on the plot, however the variations in scaling make it arduous to match throughout axes. Evaluate the orders of magnitude of proline and hue, for instance: proline has a robust optical dominance, simply due to scaling. An unscaled plot appears virtually meaningless, or at the very least very tough to interpret. Even so, faint bundles over lessons appear to look, so let’s take this as a promise for what’s but to come back…
It’s all about Scale
A lot of you (everybody?) are aware of the min-max scaling from ML preprocessing pipelines. So let’s not use that. I’ll do some standardization of the info, i.e., we do Z-scaling right here (every function can have a imply of zero and unit variance), to present all axes the identical weight.
from sklearn.preprocessing import StandardScaler
# Separate options and goal
options = df.drop("Class", axis=1)
scaler = StandardScaler()
scaled = scaler.fit_transform(options)
# Reconstruct a DataFrame with scaled options
scaled_df = pd.DataFrame(scaled, columns=options.columns)
scaled_df["Class"] = df["Class"]
plt.determine(figsize=(12,6))
parallel_coordinates(scaled_df, 'Class', colormap='plasma', alpha=0.5)
plt.title("Parallel Coordinates Plot of Wine Dataset (Scaled)")
plt.xticks(rotation=45)
plt.present()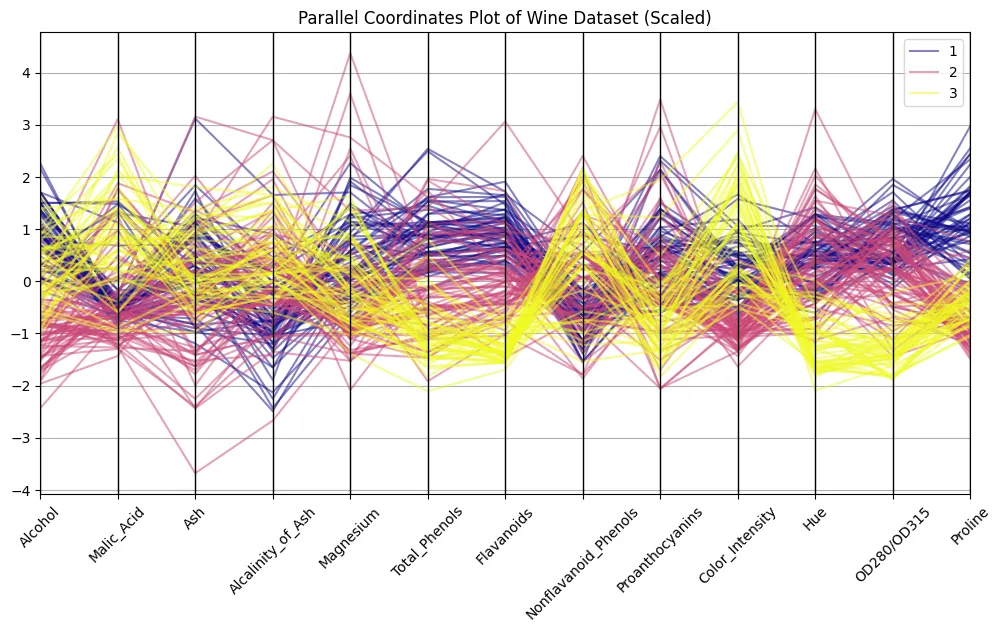
Bear in mind the image from above? The distinction is putting, eh? Now we are able to discern patterns. Attempt to distinguish clusters of strains related to every wine class to seek out out what options are most distinguishable.
Function Choice
Did you uncover one thing? Appropriate! I bought the impression that alcohol, flavonoids, color depth, and proline present virtually textbook-style patterns. Let’s filter for these and attempt to see if a curation of options helps make our observations much more putting.
chosen = ["Alcohol", "Flavanoids", "Color_Intensity", "Proline", "Class"]
plt.determine(figsize=(10,6))
parallel_coordinates(scaled_df[selected], 'Class', colormap='coolwarm', alpha=0.6)
plt.title("Parallel Coordinates Plot of Chosen Options")
plt.xticks(rotation=45)
plt.present()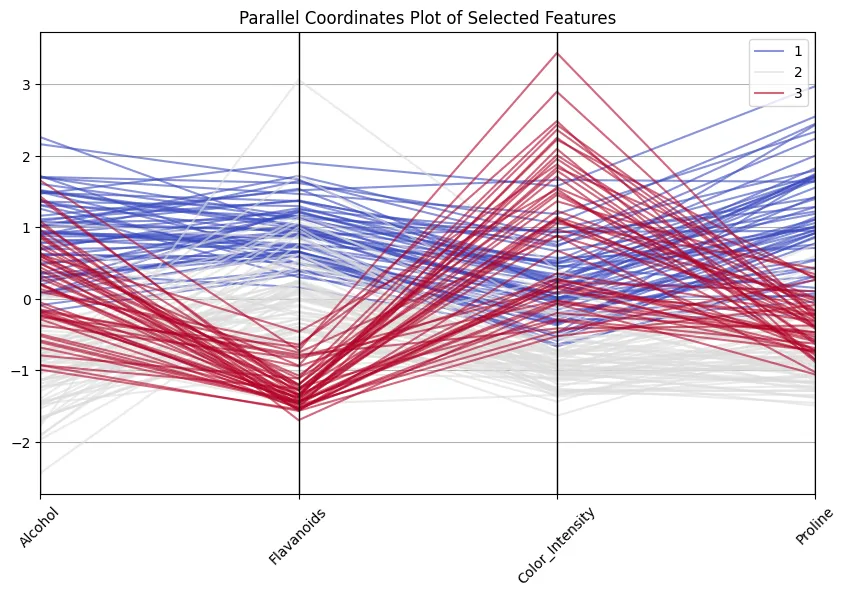
Good to see how class 1 wines at all times rating excessive on flavonoids and proline, whereas class 3 wines are decrease on these however excessive in color depth! And don’t suppose that’s a useless train… 13 dimensions are nonetheless okay to deal with and to examine, however I’ve encountered circumstances with 100+ dimensions, making lowering dimensions crucial.
Including Interplay
I admit: the examples above are fairly mechanistic. When writing the article, I additionally positioned hue subsequent to alcohol, which made my properly proven lessons collapse; so I moved color depth subsequent to flavonoids, and that helped. However my goal right here was to not provide the excellent copy-paste piece of code; it was relatively to point out you using parallel coordinates primarily based on some easy examples. In actual life, I’d arrange a extra explorative frontend. Plotly parallel coordinates, as an illustration, include a “brushing” function: there you may choose a subsection of an axis and all polylines falling inside that subset might be highlighted.
You may also reorder axes by easy drag and drop, which frequently helps reveal correlations that had been hidden within the default order. Trace: Strive adjoining axes that you just suspect to co-vary.
And even higher: scaling isn’t vital for inspecting the info with plotly: the axes are robotically scaled to the min- and max values of every dimension.
Right here’s a code so that you can reproduce in your Colab:
import plotly.categorical as px
# Hold class as a separate column; Plotly's parcoords expects numeric color for 'coloration'
df["Class"] = df["Class"].astype(int)
fig_all = px.parallel_coordinates(
df,
coloration="Class", # numeric color mapping (1..3)
dimensions=options.columns,
labels={c: c.substitute("_", " ") for c in scaled_df.columns},
)
fig_all.update_layout(
title="Interactive Parallel Coordinates — All 13 Options"
)
# The file beneath may be opened in any browser or embedded by way of <iframe>.
fig_all.write_html("parallel_coordinates_all_features.html",
include_plotlyjs="cdn", full_html=True)
print("Saved:")
print(" - parallel_coordinates_all_features.html")
# present figures inline
fig_all.present()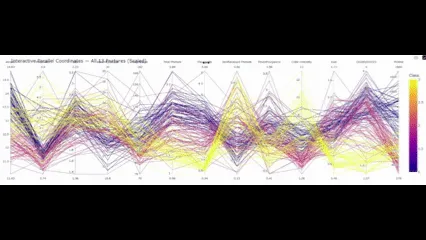
So with this ultimate ingredient in place, what conclusions will we draw?
Conclusion
Parallel coordinates usually are not a lot concerning the arduous numbers, however rather more concerning the patterns that emerge from these numbers. Within the Wine dataset, you would observe a number of such patterns – with out operating correlations, doing PCA, or scatter matrices. Flavonoids strongly assist distinguish class 1 from the others. Color depth and hue separate lessons 2 and three. Proline additional reinforces that. What follows from there’s not solely which you could visually separate these lessons, but in addition that it provides you an intuitive understanding of what separates cultivars in follow.
And that is precisely the energy over t-SNE, PCA, and so forth., these methods undertaking knowledge into parts which are glorious in distinguishing the lessons… However good luck attempting to elucidate to a chemist what “part one” means to him.
Don’t get me improper: parallel coordinates usually are not the Swiss military knife of EDA. You want stakeholders with an excellent grasp of information to have the ability to use parallel coordinates to speak with them (else proceed utilizing boxplots and bar charts!). However for you (and me) as an information scientist, parallel coordinates are the microscope you have got at all times been eager for.
Often Requested Questions
A. Parallel coordinates are primarily used for exploratory evaluation of high-dimensional datasets. They assist you to spot clusters, correlations, and outliers whereas protecting the unique variables interpretable.
A. With out scaling, options with massive numeric ranges dominate the plot. Standardising every function to imply zero and unit variance ensures that each axis contributes equally to the visible sample.
A. PCA and t-SNE scale back dimensionality, however the axes lose their unique that means. Parallel coordinates preserve the semantic hyperlink to the variables, at the price of some muddle and potential overplotting.
Login to proceed studying and revel in expert-curated content material.

